Cohort identification template
Gold standard using manual annotations created by two clinicians at the UH CMC EMU. The two clinicians annotated the 104 discharge summary reports processed by EpiDEA according to the cohort identification template.
The template was implemented as a table, which was filled by each of the clinicians after reviewing each report.
For “Current Antiepileptic Medications” and “Past Antiepileptic Medications,” only drug brand names or drug ingredients were recorded. To measure the quality of annotations, we calculated the inter-annotator agreement by manually comparing the two sets of annotations.
The partial variation in the EEG signal pattern was due to use of terms at different levels of specialization, namely clinician 1 annotated a signal pattern as “Normal,” whereas clinician 2 annotated the same signal pattern as “Sharp transients” (a sub category of “Normal”).
A total of 17,614 sentences were detected in the discharge summaries, including 118,786 word tokens and 27,899 noun phrases, which were mapped to appropriate EpSO classes.
Table 3 lists the top 8 EpSO ontology classes occurring in the discharge summaries ordered by the number of extractions.
| Ontology Class | Number of Extractions |
|---|---|
| PhysicialPathologicalProcess | 3219 |
| Seizure | 3195 |
| Organism | 968 |
| ClinicalDrugComponent | 957 |
| EpilepticSeizure | 713 |
| EEGPattern | 658 |
| AbnormalEEGpattern | 625 |
| EpileptiformPattern | 611 |
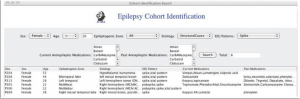
The EpiDEA visual query interface for cohort identification
To support these patient cohort identification queries over data extracted from the patient discharge summaries, we implemented a visual query interface to allow investigators to directly query the PRISM patient knowledge base
he query interface consists of a set of drop-down menus that are directly populated with the EpSO classes. This allows users to flexibly construct queries, for example they can use either the drug ingredients or drug brand. For example, “CarBAMazepine” has three trade names “Carbatrol,” “TEGretol,” and “TEGretolXR,” so in the case a user selects “CarBAMazepine,” the query interface identifies all discharge summaries that mention the drug ingredient or its tradenames.
The results of a query include the patient discharge summaries and the actual values in the discharge summaries, which allow investigators to manually verify the result values.
The query interface has been implemented using Java 1.6 and is integrated with EpiDEA system, which provides an integrated environment for investigators in the PRISM project for cohort identification.
[1]