HITEx
An open source, reusable, component-based NLP system called HITEx.
Free text clinical records contain a large amount of useful information, and NLP has the potential to unlock this wealth of information. One challenge with medical NLP tools is that they are not easy to adapt, generalize and reuse.
NLP tool that we refer to as the Health Information Text Extraction (HITEx) tool. Following the example of GATE (General Architecture for Text Engineering) [14] and using GATE as a platform, a suite of open-source NLP modules were adapted or created. We then assembled these modules into pipelines for different tasks.
HITEx uses GATE as the development platform. GATE is an open-source natural language processing framework; it includes a set of NLP modules, collectively known as a Collection of Reusable Objects for Language Engineering (CREOLE) [14]. CREOLE contains NLP modules that perform some common tasks, such as tokenizing, part-of-speech (POS) tagging, and noun phrases parsing.
The GATE framework can be viewed as a backplane for plugging in CREOLE components. The framework provides various services to the components, such as component discovery, bootstrapping, loading and reloading, management and visualization of data structures, and data storage and process execution. GATE is an active project at the University of Sheffield, UK with a large user community worldwide.
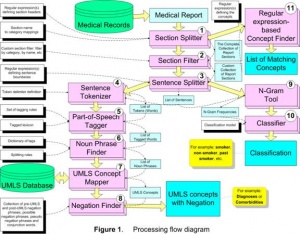
HITEx uses 11 GATE modules (components), two of which were adapted from the CREOLE, and rest were developed specifically for HITEx (see Figure Figure11):
1. Section splitter splits the medical report into the sections; assigns the section to the category (categories), which are based on the section headers for each type of the medical document. For the parsing of discharge summaries and outpatient notes, we collected and categorized over 1000 section headers. For example, "principal diagnosis" is categorized as "Primary Diagnosis", while the "discharge medications" header is mapped to both the Discharge and Medications categories. The header collections are not part of the section splitter, but supplied to the splitter as a configuration file.
2. Section filter selects the subset of sections based on the selection criteria, such as category name, section name, etc. This module uses a simple expression language that allows rather complex criteria expressions to be created.
3. Sentence splitter splits the section into sentences. The module relies on the set of regular expression-based rules that define sentence breaks.
4. Sentence tokenizer splits the sentence into the tokens (words). The module uses the extensive set of regular expressions that define both token delimiters and special cases when certain punctuation symbols should not be used as token delimiters (e.g. decimal point in numbers, period in some multi-word abbreviations, etc).
5. POS tagger assigns part-of-speech tags to each word (token) in the sentence. This module is based on the Brill-style, rule-based POS tagger, originally written by Mark Hepple [16] as a plug-in for the Gate framework.
6. Noun phrase finder groups POS-tagged words into the noun phrases using the set of rules and the lexicon. This module is an implementation of the Ramshaw and Marcus transformational learning-based noun phrase chunker [17]. The original version is available as a Gate framework plug-in.
7. UMLS concept mapper maps the strings of text to UMLS (Unified Medical Language System) concepts. The module first attempts exact match; when exact matches are not found, it stems, normalizes and truncates the string. For instance, "failures of heart" is mapped to the concept "Heart Failure" and "back pain and asthma" is mapped to concepts "Back Pain" and "Asthma".
8. Negation finder assigns the negation modifier to the existing UMLS concepts. Currently, this module is an implementation of NexEx-2 negation algorithm developed by Chapman et al. [18].
9. N-gram tool extract n-word text fragments along with their frequency from a collection of text.
10. Classifier takes a smoking-related sentence to determine the smoking status of a patient. The classifier is a support vector machine (SVM) using single words as features. It was trained and tested on a data set of about 8500 smoking-related sentences through 10-fold cross-validation. To create the classifier, we experimented with naïve Bayes, SVM and decision trees, and 1–3 word phrase features using Weka [19], which is a publicly available tool kit. The detailed description of the experiments can be found in a previous publication [15].
11. Regular expression-based concept finder finds all occurrences of the concepts defined as a regular expression in the input chunk of text. For example: medications, smoking keywords, etc.
Each module expects a set of parameters/configuration files. Take the task of extracting principal diagnosis for example, we would specify the primary diagnosis headers for the Section Filter module. For each task, a different pipeline of modules may be assembled. To extract diagnoses, Section Splitter, Section Filter, Sentence Splitter, Sentence Tokenizer, Part-of-Speech, Noun Phrase Finder, UMLS Concept Mapper and Negation Finder modules would be applied sequentially. While for extracting smoking status, Section Splitter, Section Filter, Sentence Splitter, N-gram tool and Classifier are formed into a pipeline.
Except the smoking classifier module, none of the modules had been formally evaluated. In ad-hoc testing, results obtained from the modules were satisfactory.
Methods
A large data set containing records on approximately 97,000 asthma and COPD patients was obtained from the Partners' Health Care Research Patient Data Repository (RPDR). The RPDR data warehouse includes a wide variety of records (not deidentified) including free text, administrative codes, laboratory codes and text, and numerous other data sources for all encounters for all patients at all Partners facilities. The patients included in our data set had one or more asthma or COPD related admission diagnosis (determined by their ICD9 billing codes) in one or more of their RPDR records. The RPDR population includes 90–98% of the total Partners patient population. The study was approved by the Brigham and Women's Hospital's institutional review board; the protocol number is 2004P002260 (Subphenotypes in Common Airways Disorders).
In the evaluation, we focused on discharge summaries, in particular, those related to hospitalization caused by asthma or COPD exacerbation. We collected a random sub-sample of 150 discharge summaries from the data set that either has had an asthma or COPD related ICD9 billing code or contains an asthma or COPD related string from an extensive list of related concepts and names that we manually identified.
An asthma expert (STW) reviewed the 150 reports and answered the following questions for each report:
1. Principal diagnosis include asthma: yes/no/insufficient data
2. Principal diagnosis include COPD: yes/no/insufficient data
3. Co-morbidities include asthma: yes/no/insufficient data
4. Co-morbidities include COPD: yes/no/insufficient data
5. Smoking status: current smoker, past smoker, non smoker, patient denies smoking, insufficient data
Similarly, HITEx was used to answer the same 5 questions for each report. First, we created three HITEx pipelines to extract principal diagnosis, co-morbidities and smoking status, respectively. The principal diagnosis and co-morbidities were then processed to determine if they contained an asthma or COPD diagnosis. To do so, we used the relationship table (MRREL) in the UMLS. All descendants of the asthma and COPD concepts were considered to be a type of asthma and COPD. Diagnoses extracted from the principal diagnosis sections (determined by the section headers) were deemed as principal. When we could not determine if a diagnosis was primary or secondary because of the lack of header information, it was considered to be primary by default.
We also used ICD9 codes to answer the questions regarding the principal diagnosis and co-morbidities. The ICD9 hierarchy was used to determine if a code is asthma or COPD related. The codes are: Asthma – 493.*; COPD – 490–492.*, 494–496.*, 466.*.
For asthma/COPD principal diagnosis and co-morbidities, we compared the HITEx and ICD9 answers to that of the human expert. For smoking status, HITEx results were compared to the human expert's.
Generally speaking, NLP programs infer "yes" for a diagnosis if certain string patterns are found, "no" if they are not found, but often do not have a notion of "insufficient data". On the other hand, the human label of "insufficient data" could result from the absence of explicit information, the presence of ambiguous information or the presence of conflicting information. To compare HITEx results to the human ratings, we treated the "insufficient data" label in three ways: exclude cases with the label, regard it as "yes", and regard it as "no". In the case of smoking, though, non-smoker status are often explicitly stated so "insufficient data" was interpreted to mean that no smoking-related information was found.
In addition, we experimented with two ways to combine ICD9 and HITEx results to improve the diagnosis extraction performance:
NLP and ICD9:
- Both are 'YES' → 'YES'
- everything else → 'NO'
NLP or ICD9:
- either one is 'YES' → 'YES'
- everything else → 'NO'
We calculated the accuracy of HITEx, ICD9 and HITEx-ICD9 combinations, i.e. the percentage of cases which HITEx, ICD9 or HITEx-ICD9 combinations agreed with the expert. Sensitivity and specificity were also calculated, assuming the human expert as the correct classifier. In practice, a human's ability to maintain focus tends to decline after a few hours of this relatively tedious chart review, so the human expert's answers were confirmed by four other physician members of the I2B2 team. Some obvious errors and omissions were corrected by a consensus of clinicians at the team meeting.
[1]