NLP Tools (I2B2 Project)
Textual medical records contain need to be extracted and/or indexed in order to be analyzed and interpreted by automated tools. A collection of natural language processing (NLP) tools to extract various types of information from unstructured medical records. Although a number of NLP systems had demonstrated good accuracy in information extraction, they were often domain-, institution- and application-specific.
The generic NLP components, when assembled in pipelines and initialized with custom configuration parameters, become a powerful medical data mining instrument. Medical concepts such as diagnoses, comorbidities, discharge medications, and smoking status can be ascertained. A textual medical record is a rich source of clinical information.
A suite of NLP tools for the I2B2 (Informatics for Integrating Biology and the Bedside, a national center for biomedical computing) project to address a wide range of text processing needs.
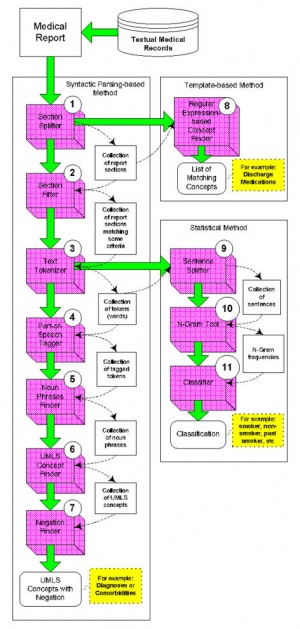
A modularized and parameterized approach in the software development and employed syntactic, statistical, template-based methods for different parsing tasks. This approach allows users to tailor the NLP tools to extract and index specific information from different domains and institutions.
Methods
I2B2 developed 11 modules for text report processing
- Section Splitter
- Section Filter
- Text Tokenizer
- Part-of-Speech (POS) Tagger
- Noun Phrase Finder
- UMLS Concept Finder
- Negation Finder
- Regular Expression-based Concept Finder
- Sentence Splitter
- N-Gram Tool
- Classifier (e.g. Smoking Status Classifier)